Features
SecureDNA was designed to screen all oligo and gene synthesis orders against an up-to-date list of known and emerging hazards, including pre-generated functional variants.
Universal - fast, free, and available to all
-
Fast
Rapid and delay-free screening at thousands of base pairs / sec, plus optional customer pre-screening
-
Free
Non-profit and free to use, thanks to several generous donations and our efficient exact-match algorithm
-
Accessible
Available to all providers with free integration support, to safeguard DNA synthesis worldwide
Comprehensive - sensitive, specific, and up-to-date
-
Sensitive
Detects all known hazards and predicted functional variants down to 30 b.p., preventing oligos too small to be screened by other methods from being used to assemble hazards
-
Specific
Outputs ~0 false alarms by using patented reverse screening to remove known non-hazards, enabling use in synthesizers and assemblers without human support
-
Up-to-date
Uses a centralized database, updated swiftly by our biosecurity team as new threats are identified; no need to update local installations
Trustworthy - privacy-preserving and neutral
-
Privacy-preserving
Protected by multiple layers of cryptography & distributed around the world, your customers’ sequences will be uninterpretable - by us or anyone else
-
Politically neutral
Run within a Swiss non-profit foundation without involvement by any government - a worldwide research collaboration
Detailed description of the platform
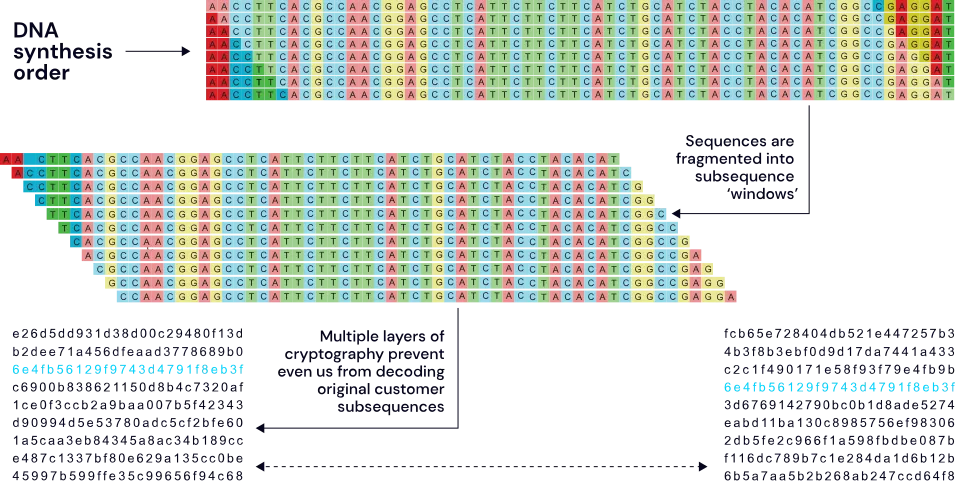
The SecureDNA system uses a novel exact-match search algorithm that finds matches to hazards and functional equivalents. It does this by generating the set of all hazard subsequences of a predefined length, choosing some to defend quasi-randomly, then using algorithms to generate millions of functionally equivalent subsequences of each. These are subjected to reverse screening by comparing them to existing sequence databases in order to remove any that match known non-hazardous entries.
The reverse screening step virtually eliminates false positives, the inclusion of quasi-random functional variants prevents adversaries from redesigning or mutating the hazard to evade screening, and searching for 30 base pair subsequences makes it very difficult to assemble hazards from oligonucleotides that are too small to be screened using other methods.
Because exact-match screening is computationally efficient, we can apply oblivious multi-party cryptography to protect the confidentiality of both orders and the hazard database. Using this scheme, each possible subsequence of an order to be screened is turned into a unique cryptographic hash, and each such hash is then looked up in the database of hazards.
This hash requires multiple separate machines to compute, meaning that even an attacker who could eavesdrop on network traffic or compromise a subset of computers in the screening system could not learn the plaintext of a customer order, and not even SecureDNA itself ever receives the customer’s DNA sequences—they remain safely on-premises at a provider or within the hardware of a benchtop synthesizer.
Screening for hazardous DNA sequences is provided as a free service by the SecureDNA Foundation, an independent nonprofit foundation in the sense of Article 80 et seqq. Swiss Civil Code with legal seat in Zug, Switzerland. The purpose of the Foundation is to develop, maintain, administrate, distribute, and encourage the universal adoption of software for screening nucleic acid sequences. The Foundation may support any action intended to prevent nucleic acids from being used to cause harm.
Partners and Support
We are grateful for support from the Open Philanthropy Project to team members at MIT, Aarhus University, and Northeastern University, and to an anonymous Chinese philanthropist for a donation to team members at Tsinghua University and Shanghai Jiao Tong University.
SecureDNA screening demo